- Location : Home» Newsroom
GhDMT7-mediated DNA methylation dynamics enhance starch and sucrose metabolism pathways to confer salt tolerance in cotton
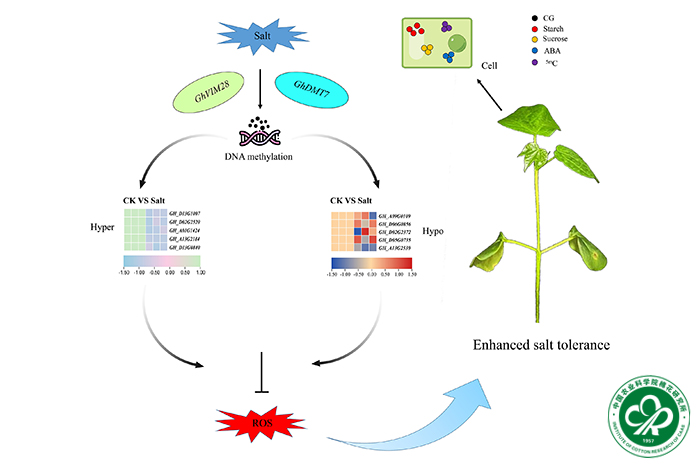
This study provides a comprehensive analysis of the impact of DNA methylation in cotton under salt stress conditions, elucidating its effects on gene expression and biological processes. Here, we determined the structures of the DNA methylation landscape across the cotton genome subjected to salt stress using whole-genome bisulfite sequencing (WGBS) and RNA-seq methodologies. We identified 4938 differentially methylated regions (DMRs) correlated with alterations in gene expression. Salt stress induced significant shifts in DNA methylation patterns, particularly in CHH contexts, suggesting context-dependent epigenetic regulation. DMRs were found to be implicated in diverse biological processes and pathways, encompassing protein metabolism, cellular homeostasis, starch and sucrose metabolism, and plant hormone signaling, all pivotal for cotton's adaptation to salt stress. Furthermore, RNA-seq analysis confirmed the impact of DNA methylation on gene expression, uncovering 9642 salt stress-responsive differentially expressed genes (DEGs). These DEGs exhibited enrichment in pathways such as carbohydrate metabolism, cell wall synthesis, and defense response, underscoring the intricate interplay between methylation and gene regulation in stress response. Moreover, the study investigated the role of the key DNA methyltransferase gene GhDMT7 in modulating cotton's response to salt stress, revealing that its downregulation enhanced cotton's salt tolerance, potentially attributed to decreased DNA methylation levels, reduced membrane damage, and enhanced antioxidant capacity. These findings elucidate the role of DNA methylation in abiotic stress resilience and provide insights for crop improvement.